
Home
>> News
December 2025
It is published!!!!!!!
Kaivalya Walavalkar, Shivani Gupta, Jelena Kresoja-Rakic, Mathieu Raingeval, Chiara Mungo, Philip Rubin & Raffaella Santoro
Single-cell dynamics of genome-nucleolus interactions captured by nucleolar laser microdissection (NoLMseq)
Nature Communications (2025) 16(1):11417
> Full Text Article

Gene positioning in nuclear space is central to regulation, with repressive chromatin domains often contacting the nuclear lamina or nucleoli. The nucleolus undergoes structural changes in different cellular states, potentially altering genome organization. Yet, how nucleolar states influence 3D-genome architecture remains underexplored, largely due to the lack of methods able to map nucleolar-associated domains (NADs) in single cells and stressed nucleoli. We developed NoLMseq, a technique combining laser-capture microdissection and DNA sequencing to map NADs in single cells. NoLMseq uncovered unexplored features of chromosome organization around nucleoli, including NAD heterogeneity in mouse embryonic stem cells, yielding two major populations with distinct chromatin and developmental states. NADs predominantly contact nucleoli monoallelically, with contact frequency correlating with gene expression and chromatin states. Under nucleolar stress, NoLMseq revealed extensive chromosome reorganization, highlighting the importance of nucleolus integrity in genome organization. Thus, NoLMseq provides a critical tool to study 3D-genome responses to nucleolar states in health and disease.
PS. This work did not deserve the unfair treatment we have to deal with such as unnecessary multiple round of revisions, editors not taking decisions, and more. Thus, we are very firm to not thank the non-anonymous Reviewer 1. The file of the peer review process here will explain everything!
In this review, we described the recent methods to study chromosome architecture around nucleoli.
The nucleolus is the largest subcompartment of the nucleus, known to be the place of ribosome biogenesis. Emerging evidence has started to implicate the nucleolus in the organization of chromosomes in the nucleus. Genomic domains contacting the nucleolus are defined as nucleolar associated domains (NADs) and are generally characterized by repressive chromatin states. However, the role of the nucleolus in genome architecture remains still not fully understood mainly because the lack of a membrane has challenged the establishment of methods for accurate identification of NADs. Here, we will discuss recent advances on methods to identify and characterize NADs, discuss their improvements relative to old methods, and highlight future perspectives.
May 2025
It is published!
Gupta, S., Bersaglieri, C., Baer, D., Raingeval, M., Schaab, L., and Santoro, R.
The nucleolar granular component mediates genome-nucleolus interactions and establishes their repressive chromatin states
Molecular Cell (2025) 85(11):2165-2175.e6

Repressive chromatin domains often localize to the nuclear lamina or nucleolus. Although nucleolar-associated domains (NADs) have recently been mapped, their mechanisms of nucleolar association and functional significance remain unclear. Here, we show that nucleophosmin (NPM1), a factor located in the granular component of the nucleolus, mediates NAD association in mouse embryonic stem cells. NPM1 binds NADs, interacts with the histone methyltransferase G9a (EHMT2), and is required for establishing H3K9me2 at NADs. Loss of NPM1 or expression of a DNA-binding-deficient mutant disrupts NAD-nucleolus association and reduces H3K9me2 specifically at NADs. G9a is dispensable for NAD-nucleolus contacts, indicating that H3K9me2 is acquired after NADs associate with NPM1 at nucleoli. These findings reveal mechanistic insights into how genomic domains associate with nucleoli and form repressive chromatin and indicate that the nucleolus not only serves as a scaffold for positioning repressive domains but also plays a direct role in establishing their repressive chromatin states.
PS. Fair reviewing process thanks to the editor and reviewers #1 and #2, but not reviewer #3.
April 2024
It is published!!!!!
Luigi Lerra, Martina Panatta, Dominik Bär, Isabella Zanini, Jennifer Yihong Tan, Agnese Pisano, Chiara Mungo, Celia Baroux, Vikram Govind Panse, Ana C. Marques, Raffaella Santoro
An RNA-dependent and phase-separated active subnuclear compartment safeguards repressive chromatin domains
Molecular Cell (2024) doi: 10.1016/j.molcel.2024.03.015
This work was a true detective story! The project started since we wanted to analyse the function of BAZ2A in mouse embryonic stem cells. It was a puzzle since the beginning ... but then the data started to make sense, and the results were very surprising and exciting.
The nucleus is composed of functionally distinct membrane-less compartments which undergo phase separation (PS). However, whether different subnuclear compartments are connected remain elusive. We identified a type of nuclear bodies with PS features composed of BAZ2A that associates with active chromatin. BAZ2A-bodies depend on RNA transcription and BAZ2A non-disordered RNA-binding TAM domain. Although BAZ2A and H3K27me3 occupancies anticorrelate in the linear genome, in the nuclear space BAZ2A-bodies contact H3K27me3-bodies. BAZ2A-bodies disruption promotes BAZ2A invasion into H3K27me3-domains, causing H3K27me3-bodies loss and gene upregulation. Weak BAZ2A-RNA interactions, such as with nascent transcripts, promote BAZ2A-bodies whereas the strong binder lncRNA Malat1 impairs them while mediating BAZ2A association to chromatin at nuclear speckles. In addition to unravelling a direct connection between nuclear active and repressive compartments through PS mechanisms, the results also showed that the strength of RNA-protein interactions regulates this process contributing to nuclear organization and the regulation of chromatin and gene expression.
PS. It took us almost 12 months to get published. However, only 2 months were spent for the revision (we had already the majority of the requested data), the rest was waiting, and waiting for the response of the Editor. 3.5 months for a decision to send the revised manuscript to Reviewers, 3 months for a decision after the revised version was back from Reviewers. Two Reviewers agreed for publication, another Reviewer asked to take away an experiment and supported the publication. Editor rejected the paper and, as she has always done, did not take a second to reply to our request of clarifications. So unprofessional!!!! Transfer to Mol Cell, very professional and supportive Editor, paper accepted without further changes.

March 2024
It is published!!!!!
Meneka Rupasinghe, Cristiana Bersaglieri, Deena M Leslie Pedrioli, Patrick G. A. Pedrioli, Martina Panatta, Michael O. Hottiger, Paolo Cinelli, Raffaella Santoro
PRAMEL7 and CUL2 decrease NuRD stability to establish ground-state pluripotency
EMBO Reports (2024) 25(3):1453-1468
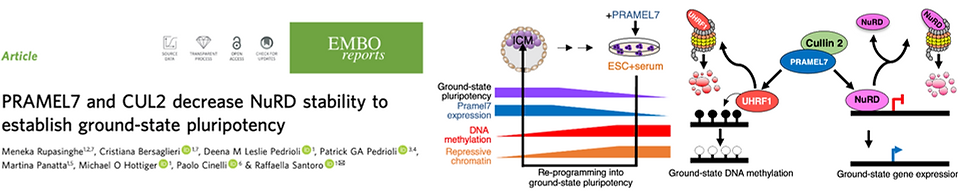
In this work, we show how Pramel7 regulates ground-state pluripotency, which is established in E4.5 preimplantation epiblast. Embryonic stem cells (ESCs) represent the immortalization of pluripotency, however, their gene expression signature only partially resembles that of developmental ground-state. Induced PRAMEL7 expression, a protein highly expressed in the ICM but lowly expressed in ESCs, reprograms developmentally advanced ESC+serum into ground-state pluripotency by inducing a gene expression signature close to developmental ground-state. However, how PRAMEL7 reprograms gene expression remains elusive. Here we show that PRAMEL7 associates with Cullin2 (CUL2) and this interaction is required to establish ground-state gene expression. PRAMEL7 recruits CUL2 to chromatin and targets regulators of repressive chromatin, including the NuRD complex, for proteasomal degradation. PRAMEL7 antagonizes NuRD-mediated repression of genes implicated in pluripotency by decreasing NuRD stability and promoter association in a CUL2-dependent manner. Our data link proteasome degradation pathways to ground-state gene expression, offering insights to generate in vitro models to reproduce the in vivo ground-state pluripotency.
PS. Strange story to publish this paper. First journal, hypercritical reviewers, revised version with all addressed points sent to the Journal, Editor decided to not send it back to Reviewers! Sent to EMBO Reports (better Journal than the other), very positive Reviewers who congratulated with us for the nice work, only minor points, mainly clarifications, paper accepted. What was changed? Just the competence and expertise of Editors and Reviewers.
May 2023
It is published!!!!!
Cristiana Bersaglieri and Raffaella Santoro
Methods for mapping 3D-chromosome architecture around nucleoli
Curr Opin Cell Biol (2023)
doi: 10.1016/j.ceb.2023.102171

April 2023
It is published!!!!!
Marcin Roganowicz, Dominik Bär, Cristiana Bersaglieri, Rossana Aprigliano, Raffaella Santoro
BAZ2A-RNA mediated association with TOP2A and KDM1A represses genes implicated in prostate cancer
Life Science Alliance (2023) DOI: 10.26508/lsa.202301950.

BAZ2A represses rRNA genes (rDNA) that are transcribed by RNA polymerase I. In prostate cancer (PCa), BAZ2A function goes beyond this role because it represses genes frequently silenced in metastatic disease. However, the mechanisms of this BAZ2A-mediated repression remain elusive. Here, we show that BAZ2A represses genes through its RNA-binding TAM domain using mechanisms differing from rDNA silencing. Although the TAM domain mediates BAZ2A recruitment to rDNA, in PCa, this is not required for BAZ2A association with target genes. Instead, the BAZ2A-TAM domain in association with RNA mediates the interaction with topoisomerase 2A (TOP2A) and histone demethylase KDM1A, whose expression positively correlates with BAZ2A levels in localized and metastatic PCa. TOP2A and KDM1A pharmacological inhibition up-regulate BAZ2A-repressed genes that are regulated by inactive enhancers bound by BAZ2A, whereas rRNA genes are not affected. Our findings showed a novel RNA-based mechanism of gene regulation in PCa. Furthermore, we determined that RNA-mediated interactions between BAZ2A and TOP2A and KDM1A repress genes critical to PCa and may prove to be useful to stratify prostate cancer risk and treatment in patients.
April 2022
It is published!!!!!
Argonaute proteins regulate a specific network of genes through KLF4 in mouse embryonic stem cells
Stem Cell Reports (2022) S2213-6711(22)00147-3


A great collaborations with Constance Ciaudo group!
The Argonaute proteins (AGOs) are well known for their role in post-transcriptional gene silencing in the microRNA (miRNA) pathway. Here we show that in mouse embryonic stem cells, AGO1&2 serve additional functions that go beyond the miRNA pathway. Through the combined deletion of both Agos, we identified a specific set of genes that are uniquely regulated by AGOs but not by the other miRNA biogenesis factors. Deletion of Ago2&1 caused a global reduction of the repressive histone mark H3K27me3 due to downregulation at protein levels of Polycomb repressive complex 2 components. By integrating chromatin accessibility, prediction of transcription factor binding sites, and chromatin immunoprecipitation sequencing data, we identified the pluripotency factor KLF4 as a key modulator of AGO1&2-regulated genes. Our findings revealed a novel axis of gene regulation that is mediated by noncanonical functions of AGO proteins that affect chromatin states and gene expression using mechanisms outside the miRNA pathway.

March 2022
It is published!!!!!
Genome-wide maps of nucleolus interactions reveal distinct layers of repressive chromatin domains
>Full Text Article
Eukaryotic chromosomes are folded into hierarchical domains, enabling the organization of the genome into functional compartments. Nuclear periphery and nucleolus are two nuclear landmarks thought to contribute to repressive chromosome architecture. However, while the role of nuclear lamina (NL) in genome organization has been well documented, the function of the nucleolus remains under-investigated due to the lack of methods for the identification of nucleolar associated domains (NADs). Here we established methodologies based on DamID and HiC that generated accurate genome-wide maps of NADs in ESCs and neural progenitors (NPCs), revealing unprecedent layers of genome compartmentalization with distinct, repressive chromatin states based on the interaction with the nucleolus, NL, or both. NADs showed higher H3K9me2 and lower H3K27me3 content than regions exclusively interacting with NL. Upon ESC differentiation into NPCs, chromosomes around the nucleolus acquire a more compact, rigid architecture with neural genes moving away from the nucleolus and becoming unlocked for later activation. Further, histone modifications and the interaction strength within A and B compartment of NADs and LADs in ESCs set the choice to associate with the NL or nucleolus upon dissociation from their respective compartments during differentiation.
The methodologies here developed will finally make possible to include the contribution of the nucleolus in nuclear space and genome function in diverse biological systems.
January 2022
We have been awarded of a 3-year collaborative project between my lab (SNSF) and the lab of Helena Fulková (GAČR) to study genome architecture of the nucleolus in early mouse embryonic development-
We are recruiting for this exciting project!

August 2021
It is published!!!!!
Epigenetic control of melanoma cell invasiveness by the stem cell factor SALL4
>Full Text Article
A great collaboration with Lukas Sommer lab at the University of Zurich showing that the embryonic stem cell transcription factor SALL4 regulates phenotype switching in melanoma through an HDAC2-mediated mechanism.
August 2021
It is published!!!!!
BAZ2A-mediated repression via H3K14ac-marked enhancers promotes prostate cancer stem cells
>Full Text Article


Prostate cancer (PCa) is one of the most prevalent cancers in men. Unfortunately, current therapeutic approaches for PCa remain insufficient for some patients with progressive disease. Targeting of the androgen receptor (AR) axis in patients with PCa relapse was shown to be efficient only for a short period of time. These treatments are not curative; a population of cells resistant to androgen-deprivation therapy emerges and PCa becomes unresponsive and progresses to a castrate-resistant prostate cancer and metastasis, with limited treatment options. Due to the enticing possibility that PCa aggressiveness and relapse arises from PCa stem cells, the development of stem cell-specific anticancer drugs constitutes an attempt to innovate in the treatment of PCa.
Here we show that BAZ2A is an epigenetic reader of H3K14ac and regulates prostate cancer stem cells though its association with inactive enhancers marked by H3K14ac.
-
The BAZ2A-bromodomain (BRD) is an epigenetic reader of H3K14ac.
-
BAZ2A associates with H3K14ac-marked inactive enhancers that repress genes frequently silenced in aggressive PCa.
-
The H3K14 acetyltransferase EP300 is required for the repression of BAZ2A-target genes in PCa cells.
-
Pharmacological inactivation of BAZ2A with BAZ2A-BRD inhibitors impairs PCa stem cells and PTEN-loss mediated oncogenic transformation using PCa organoids.
Our findings indicate a role of BAZ2A-BRD in PCa stem cell features and suggest potential epigenetic-reader therapeutic strategies to target BAZ2A in aggressive PCa.
June 2021
Congratulations to Kaivalya for being awarded with the EMBO postdoctoral fellowship!

June 2021
Congratulations to Valdemar for being awarded with the Zurich KrebsLiga grant!

Valdemar will work on a collaborative project with Jean-Pierre Bourquin group at the University Hospital Zurich and our group that aims to elucidate the molecular mechanisms of the transcriptional circuitries controlled by the the chimeric fusion transcription factor TCF3-HLF that drives a lethal leukemia with fatal outcome.
May 2021
It is finally out!
Open access Introduction to Epigenetics.
After only 2 months >21K downloads!
This book was inspired by the course “Epigenetics” at ETH Zurich that myself together with Renato Paro, Ueli Grossniklaus, and Anton Wutz are carrying on.
The first chapters provide an introduction to the biology of chromatin with all the components and cellular processes that establish the toolbox of epigenetic mechanisms. In the following chapters, complex epigenetic phenomena are illustrated by explaining the structures and principles of the underlying molecular mechanisms. Towards the end of the book, two chapters examine environmental influences on epigenetic control and epigenetic misregulation in human disease.
October 2020
Finally out!!!! BAZ2A safeguards genome architecture of ground-state pluripotent stem cells.
>Full Text Article
Our work showing how genome architecture is regulated in ground-state ESCs is published in EMBOJ! From now on we are using the official name of TIP5... Baz2a!

How genome organization is regulated in embryonic stem cells (ESC) according to cell state and chromatin structure remains elusive. Here, the systematic comparison between ground-state mouse ESCs (ESC+2i) and developmentally advanced ESCs (ESC+serum) reveals that Baz2a (TIP5) is a critical regulator of genome partitioning and cell identity.
-
BAZ2A binds at active and open genome and supports growth of ground-state but not advanced ESCs.
-
BAZ2A interacts with SNF2H, TOP2A and cohesin on ESC chromatin.
-
BAZ2A associates with chromatin sub-domains within the active A-compartment that intersect through long-range contacts.
-
BAZ2A deletion perturbs gene expression and histone H3K27 trimethylation in ground-state ESCs.
-
BAZ2A limits invasion of ESC active chromatin into inactive domains.
June 2020
Congratulations to Marcin for being awarded with the prestigious Forschungskredit of the University of Zurich!


February 2020
Our work on prostate cancer is published in PNAS!
TIP5 primes prostate luminal cells for the oncogenic transformation mediated by PTEN-loss
We modeled prostate cancer initiation using organoids and showed that TIP5 (BAZ2A) is critical for the initiation of PCa of luminal origin mediated by Pten-loss whereas it is dispensable once Pten-loss mediated transformation is established. The order of events matters! Cross-species transcriptomic analyses (patients and murine prostate cancer organoids) revealed a PTEN-loss gene signature that identified a set of aggressive tumors with PTEN deletion, or low PTEN expression, and high-TIP5 expression. This work provides a powerful tool to elucidate prostate cancer mechanisms.
October 2019
Our review on the regulation of the nucleolus and rRNA gene chromatin during ealy development is now published in Trends in Genetics with a dedicated cover.
Nucleolus and rRNA Gene Chromatin in Early Embryo Development

September 2019
Congrats to Cristiana, who won the prize for best PhD poster for her work on the identification of nucleolar associated domains (NADs) at the Epigenesys Conference in London!
August 2019
Congratulations to Cristiana and her group for the best project proposal at the 3rd NCCR RNA & Disease Summer School (August 26th-30th 2019, Saas-Fee prize)!

July 2019
Our review in Trends in Genetics “Nucleolus and rRNA Gene Chromatin in Early Embryo Development” is published online
June 24, 2019
Congratulations to Karolina for her successful PhD defense!

June 2019
Our review in Cells “Genome Organization in and around the Nucleolus” is published
May 2019
We have been awarded a collaborative grant from the Cancer Research Center Zurich. This collaboration between Jean-Pierre Bourquin group at the University Hospital Zurich and our group aims to elucidate the molecular mechanisms of the transcriptional circuitries controlled by the the chimeric fusion transcription factor TCF3-HLF that drives a lethal leukemia with fatal outcome.

May 20, 2019
Congratulations to Rodrigo for his successful PhD defense!

May 2019
Congratulations to Rodrigo for the “Poster prize" at the 1st Comprehensive Cancer Center Zürich Scientific Retreat 2019
"Pharmacological targeting of the epigenetic reader TIP5-bromodomain impairs prostate cancer stem cells”

March 2019
Our second joint lab retreat with the groups of Constance Ciaudo (ETH Zurich) and Ana Marques (University of Lausanne). 3 days full of great science and fun in Diemtigtal, Berner Oberland


January 2019
Registration for the 3rd NCCR RNA & Disease Summer School is now open
Raffaella Santoro, Constance Ciaudo (ETH Zurich) and Ana Claudia Marques (University of Lausanne) will organize this year the third NCCR RNA & Disease Summer School on "RNA Regulation in Health and Disease: Genome architecture and gene expression - RNA turnover - Epitranscriptomics - Phase separation"
This event will take place in Saas-Fee, Switzerland from August 26th to 30th 2019.
The Summer School provides the opportunity to share ideas and learn from leading scientists in the field - check out the stunning lineup of speakers on the flyer below. The program consists of keynote presentations by invited speakers, workshops in small groups, group discussions lead by the invited speakers, flash talks by participating students and postdocs and plenty of opportunities for networking.
Click here for more information about the course and registration






